Submission of jobs is temporarily unavailable.
Due to a cluster maintenance, submission of conversion and structure calculation jobs is currently unavailable. Please check back soon. We apologize for the inconvenience.
Due to a cluster maintenance, submission of conversion and structure calculation jobs is currently unavailable. Please check back soon. We apologize for the inconvenience.
ARIA (Ambiguous Restraints for Iterative Assignment) is a software for automated NOE assignment and NMR structure calculation. It speeds up and automatizes the assignment process through the use of an iterative structure calculation scheme. Additionally, a refinement in explicit water improves the quality of the calculated structures, validation tests help spectroscopists to judge the quality of the final structures, and the support of the CCPN data model simplifies the exchange of information with other NMR software packages.
More information about ARIA can be found on the ARIA website.
The ARIAweb.pasteur.fr server provides access to the main ARIA software functionalities (NMR data conversion and structure calculation). In addition, structure calculation results can be easily visualised, with a dedicated molecular viewer and graphical displays of various validation statistics.
Access to the ARIAweb server is free. Users are encouraged to register (Signup now button on the login page) to easily manage your ARIA projects. Use of the ARIAweb server without registration is possible as an anonymous user. Still, anonymous users have the possibility to become fully registered using Create a full account in the User menu .
If you use this webserver, please cite:
Allain F, Mareuil F, Ménager H, Nilges M, Bardiaux B. ARIAweb: a server for automated NMR structure calculation. Nucleic Acids Research. 2020 Jul 2;48(W1):W41-W47. https://doi.org/10.1093/nar/gkaa362
Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998 Sep 1;54(Pt 5):905-21. https://doi.org/10.1107/S0907444998003254
Calculations performed on the ARIAweb server are organized as Projects. Users can view a list of their projects or create a new project from the their homepage. To create a new projects, use the Create a new Project button on the homepage or use the button on the projects list page. On the project creation page, enter a name for your project. If you plan to use NMR data from CCPN project archive (version2 only), upload it and click the Save button. For any project, ARIAweb offers three services:
The minimal input data to perform structure calculation using NMR data with ARIAweb are:
If those data are not already converted in the ARIA XML format, you have to first use the Data Conversion functionality (see below). Once the data are in the correct XML format, user can setup a new structure calculation (either by uploading XML file directly from their computer or by creating a new structure calculation from the data converted on the ARIAweb server (see below).
Alternatively, if your molecular and NMR data are already present in a CCPN project archive (version2 only) that you uploaded at the project creation, you can directly create a new structure calculation. CCPN project archive can be prepared from within CcpNmr Analysis (see the CcpNmr Analysis documentation for details).
To convert molecular sequences and NMR data into the ARIA XML format (and later perform structure calculation), users must first create a new Data Conversion using the New button on the project page or use the button on the list of data conversion).
Users will be asked to enter a name for their conversion project and a name for the molecule. Next, for every chain in the molecule, use the Add a chain or button. The sequence file must be uploaded in the Input field. Accepted formats are SEQ (Xeasy 3 letters format) or PDB (Protein Data Bank). See the Example section for file format.
Similarly, users can create as many NOE Spectrum as needed using the Add a Spectrum or button. Two mandatory input files are required:
Click on a format name to view an excerpt of a correctly formatted file. Input files are automatically checked for format correctness. See the Example section for example files.
When all input data have been entered, click the Finish button. You will be redirected to the list of conversions for your project. To start the data conversion process, use the Start conversion button. You will be then redirected to the Results page for your project where you can follow the status of your conversion job (see the Job status section for explanations on the different status icons). Finished jobs will be listed here. Registered users will receive a-email upon job submission with a link to the results page.
When a conversion job is finished, users can:
Need more details on ARIA conversion ? Go the ARIA documentation.
This process is identical to the use GUI of the ARIA standalone program. Users must first specified some parameters such as input data, NOE assignment criteria, number of structures to generate or parameters for the water refinement. When initiating a structure calculation from a previous conversion job on ARIAweb, mandatory input data are already pre-filled and shown next the Currently: showing the name of the file currently selected. When using data from a CCPN project, a list of possible data entries are shown for each appropriate input data type. See the Example section for file format.
The Structure Calculation form on ARIAweb is designed to guide the user through the main categories of parameters that need to be checked (if default values are used) or specified (if a user wants to change with customized values). The 4 main categories of parameters are:
In the DATA category, accepted data format for Molecule, Cross-peaks and Chemical shifts are: XML or CCPN. See the Example section for file format. Additional input data consists in restraints files formatted in CNS TBL format (or CCPN for Hydrogen bonds, distances and dihedral angle restraints). See the Example section for file format and the CNS documentation for more details.
In each category, classes of parameters are listed on the left menu. To show the corresponding form, activate it using the slider . The slider color code is as follows: inactive, current, validated, not validated.
By default, only basic options are displayed in each form. Enable the Expert Mode to see more options (User menu ).
To navigate between categories, use the Back or Next buttons below each form (this will validate the form and save what you have just entered). When you attain the end of the form and all categories icons are green, click the Save button. You will be redirected to the list of structure calculations for your project. To start the structure calculation process, use the Start ARIA job button. You will be then redirected to the Results page for your project where you can follow the status of your structure calculation job (see the Job status section for explanations on the different status icons). Finished jobs will be listed here. Registered users will receive a-email upon job submission with a link to the results page.
When a structure job is finished, users can:
Need more details on ARIA structure calculation ? Read the ARIA documentation.
As mentioned above, results for data conversion and structure calculation jobs are listed in the Results page of a project. The next figure is a screenshot of a typical Results page
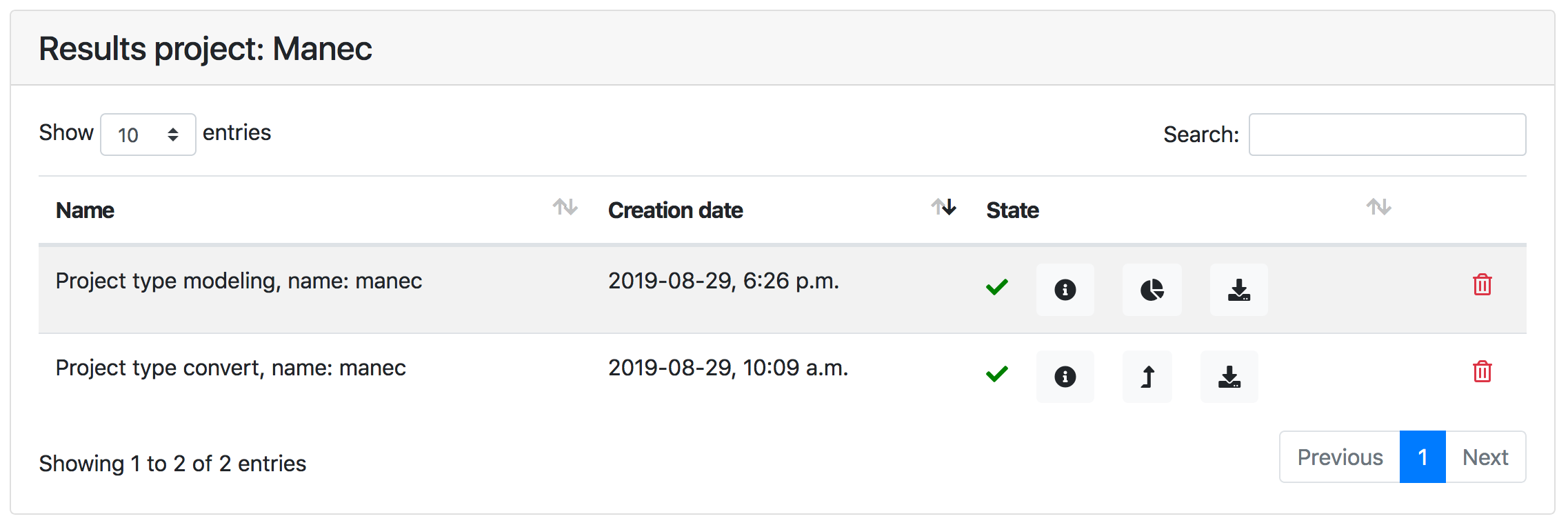
The status of a job is indicated as follows:
| Icon | Status |
|---|---|
| Building (data are uploading to the cluster) | |
| Pending (waiting to be submitted on the cluster) | |
| Running (job is running on the cluster) | |
| Success (job terminated without error) | |
| Error (job terminated with an error, check the log ) |
Results archive can be downloaded using .
For structure calculation jobs, clicking the button will show the Visualisation for the job.
The visualisation page allows to view an analysed the results of a structure calculation job. Final structure ensemble generated by ARIAweb are displayed interactively, with various representations. Final restraints statistics, structure quality checks and bundle RMSD are shown to help the user interprets the reliability of the results. On top of that, more graphs, restraints validation and PDB files can be downloaded directly.
Click here to view a live demo of the Visualisation page for structure calculation results.
The next figure is a screenshot of a typical Visualisation page, where main components are marked in red.
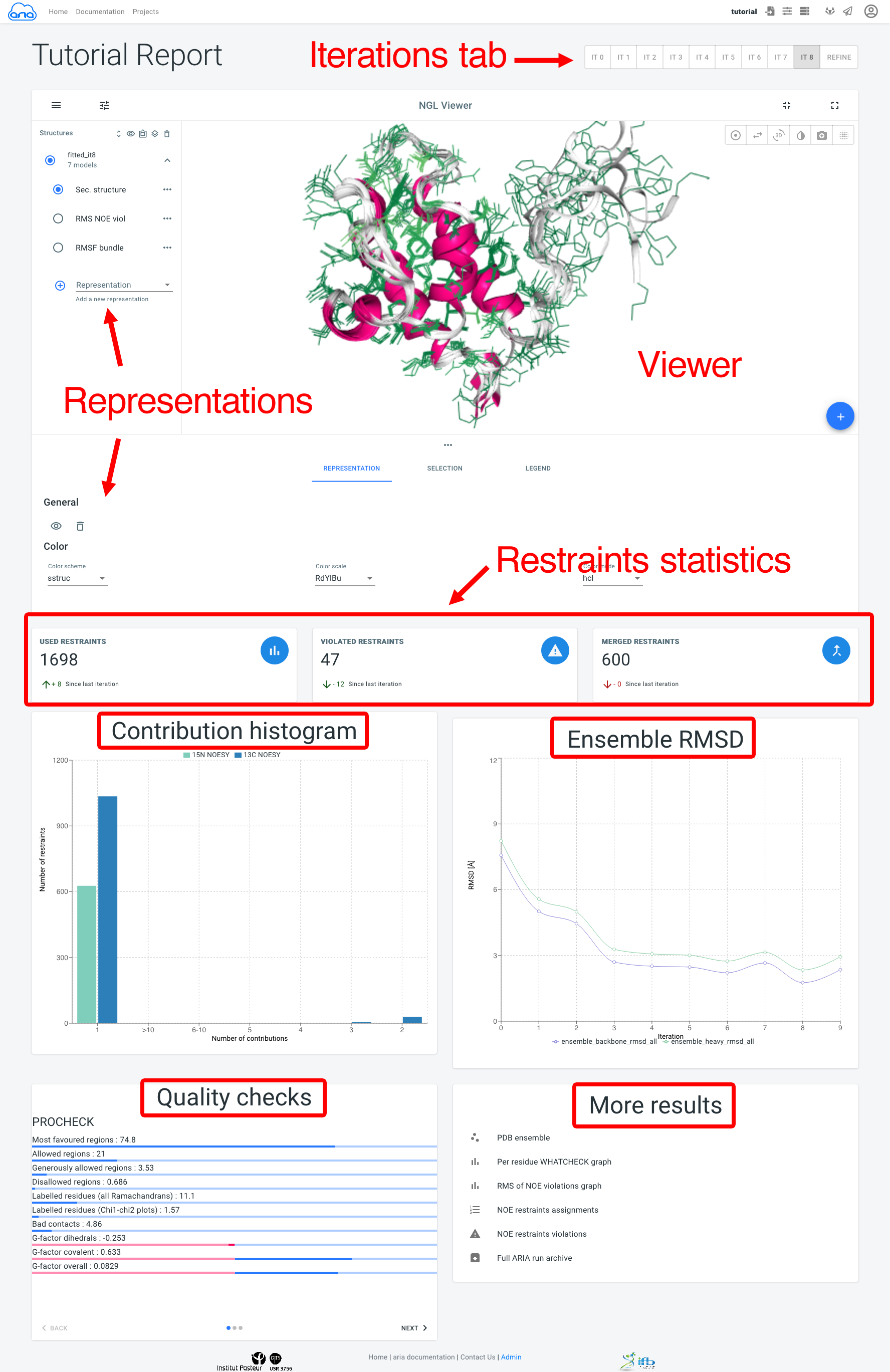
Results for all ARIA iterations can be shown by selecting an iteration in the Iterations tab. The main component of the Visualisation page is the NGL viewer. By default, several representations of the structure ensemble generated by ARIA are shown:
The Representation tab (below the NGL viewer) allows to change the visibility, styling and coloring of a selected representation. By default, all structures in the bundle are shown; use the filter_frames button to select individual models for display).
The add_circle button allows to upload another PDB file to superimpose on the ARIA structure ensemble (use the layers button to superimpose an a selected structure).
Below, Restraints statistics recapitulate the number of restraints and the trend since the previous iteration for:
The Contribution histogram gives count of restraints (from each input spectra) that have 1 (i.e. unambiguous) or more (i.e. ambiguous) assignment possibilities (or "contributions"). Ultimately, ARIA tries to reduce the ambiguity in NOE assignments, producing more unambiguous restraints. Truly ambiguous assignments can remain due to spectral overlap or chemical shifts degeneracy.
The Ensemble RMSD graph shows, for each iteration, the RMSD of the structure ensemble generated by ARIA ("bundle"), computed as the mean RMSD when superimposing of the ensemble average (after iterative superimposition). Low RMSD (< 1-2 Å) is good indicator of convergence of the NOE assignment and structure calculation process by ARIA.
The Quality checks panel summarizes the results of 3 main structural quality validators: WHAT-IF, Procheck and Molprobity. Quality scores are shown on a slider from bad to good values. Bad quality score values indicate that the input data may contain errors/inconsistencies and that ARIA was not able to produce a high quality model. We provide here some indicators on how to judge the quality checks:
Procheck Ramachandran percentage: for typical NMR structures deposited in the PDB, 80% of the dihedral angles lie within the preferred region of the Ramachandran plot. For high-resolution NMR structures, a higher percentage is expected (90%).
WHAT-IF Z-scores: WHAT-IF results are presented in the form of overall Z-scores. In general, structures with Z-scores between -2 and +2 are considered to be within a normal range and are thus good structures, while structures with Z-scores lower than -2 should be inspected further. Useful indicators of good quality are Backbone conformation and Packing quality. The bump-score also reports the number of van der Waals violations per 100 residues.
WHAT-IF profiles: recently, some studies have stressed that global structural indicators are not sufficient to detect errors in structures and suggested examining parameters on a per-residue basis. Such profiles for the WHAT-IF scores are produced by ARIA in the form of a PDF file. Thus, poor quality regions can be precisely identified.
Molprobity clashscore. this reports the number of overlaps >0.4Å per thousand atoms. For typical NMR structures deposited in the PDB, this score is generally high (>10). From our experience, the application of the log-harmonic potential along with automated weight estimation significantly improves this situation (see the More results panels).
The More results panel provides links to:
We invite users to read the following book chapter to learn more about ARIA and on how to judge the quality and reliability of structures determined with ARIA from NMR data.
Bardiaux B, Malliavin T, Nilges M. ARIA for solution and solid-state NMR. Methods Mol Biol. 2012;831:453-83. https://doi.org/10.1007/978-1-61779-480-3_23
In the demo, we will calculate the structure of the HRDC domain using 2 NOE crosspeaks lists.
To perform the demo:
Click on in your homepage
Give a name to your project (e.g. demo) and click
In the Data conversion panel, click on to automatically load sample data (sequence and cross-peaks/chemical shifts lists)
Click on to save your conversion project.
In the Data Conversion projects list, click on to submit your Data Conversion job.
Wait until the end of the job (job status )
Click on to start a Structure calculation with the converted data.
Give a name to your Structure calculation project (e.g. demo) and click on
Click on until the last form, then click to save your Structure calculation project.
In the Structure Calculation projects list, click on to submit your Structure Calculation.
Wait until the end of the job (job status )
When the job is finished, click on to start the job analysis and visualize the results.
Click here to view a live demo of the Visualisation page for Structure Calculation results on the HRDC domain.
Please follow our tutorials on how to use ARIAweb using various input data types (Xeasy, CCPN, NEF) or with an AlphaFold model as template.
https://aria.pasteur.fr/tutorial
| Tool | Data type | Format | File |
|---|---|---|---|
| Project | CCPN project | tgz | ccpn_example.tgz |
| Project | NEF file | NEF | tudor.nef |
| Conversion | Sequence | SEQ | seq_example.seq |
| Conversion | Cross-peaks list | XEasy | noesy_xeasy.peaks |
| Conversion | Chemical shifts list | XEasy | shifts_xeasy.prot |
| Structure calculation | Molecule | ARIA XML | sequence.xml |
| Structure calculation | Cross-peaks list | ARIA XML | noesy_peaks.xml |
| Structure calculation | Chemical shifts list | ARIA XML | noesy_shifts.xml |
| Structure calculation | Unambiguous distances restraints | TBL | unambig_example.tbl |
| Structure calculation | Ambiguous distances restraints | TBL | ambig_example.tbl |
| Structure calculation | H-Bond restraints | TBL | hbonds_example.tbl |
| Structure calculation | Dihedral angle restraints | TBL | dihedrals_example.tbl |
| Structure calculation | Scalar couplings restraints | TBL | karplus_example.tbl |
| Structure calculation | RDC restraints | TBL | rdcs_example.tbl |
| Structure calculation | Disulfide bridge restraints | TBL | ssbonds_example.tbl |
| Structure calculation | DNA planarity restraints | TBL | planarity_example.tbl |
| Structure calculation | Initial structure ensemble | PDB | template_iupac.pdb |
| Structure calculation | Initial Structure For Minimization | PDB | werner_iupac.pdb |
Detailed documentation about the methods and usage of the ARIA software can be found on the ARIA website.
We invite users to send enquiries specific to the use the ARIAweb server using the Contact form or by e-mail to ariaweb[at]pasteur.fr.
Questions about the ARIA software must be sent to the ARIA discussion group.
| OS | Version | Chrome | Firefox | MS Edge | Safari |
|---|---|---|---|---|---|
| MacOS | 10.14 | 78.0 | 70.0 | n/a | 12.1 |
| Linux | CentOS 7 | 78.0 | 70.0 | n/a | n/a |
| Windows | 10 | 78.0 | 70.0 | 44.18362 | n/a |
ARIAweb works together with the Institut Pasteur Galaxy instance to:
Typical execution times for Data conversion and Structure calculation jobs are around 5 min and 90 min, respectively.
Note: Execution time of jobs submitted via ARIAweb depends on the computing cluster load and the number of Pending/Running jobs as shown on the users's homepage.
ARIAweb is developed and maintained by F. Allain, F. Mareuil, T. Huynh and B. Bardiaux from the Structural Bioinformatics Unit and Bioinformatics and Biostatistics HUB at Institut Pasteur, Paris.
The development of ARIAweb is supported by the French Institute of Bioinformatics (IFB).
 **ARIA is part of the ELIXIR infrastructure. ARIA is an Elixir service.** [Read more](https://www.elixir-europe.org/about-us/why-needed).
**ARIA is part of the ELIXIR infrastructure. ARIA is an Elixir service.** [Read more](https://www.elixir-europe.org/about-us/why-needed).